Immunomics in Allergies
It is shown that around 30% of the world's population suffers from allergies at all ages. The continuing prevalence of allergy is associated with environmental and lifestyle changes. However, our understanding of the genetic determinants, the environmental determinants, and the potential interactions between these factors remains unclear. The development of high-throughput and large-scale studies based on immunomics has facilitated our understanding of the heterogeneity and complexity of allergy.
Introduction to Allergies
Allergies are the body's overreaction to allergens. Immune reactions can be IgE-mediated, non-IgE-mediated, and mixed. Following the sensitization phase, subsequent exposure to the allergen affects immune effector cells and the antigen attaches to IgE molecules that bind to FcεRI receptors on the surface of mast cells and basophils, leading to their degranulation and the release of histamine and other inflammatory mediators in the immediate allergic reaction.
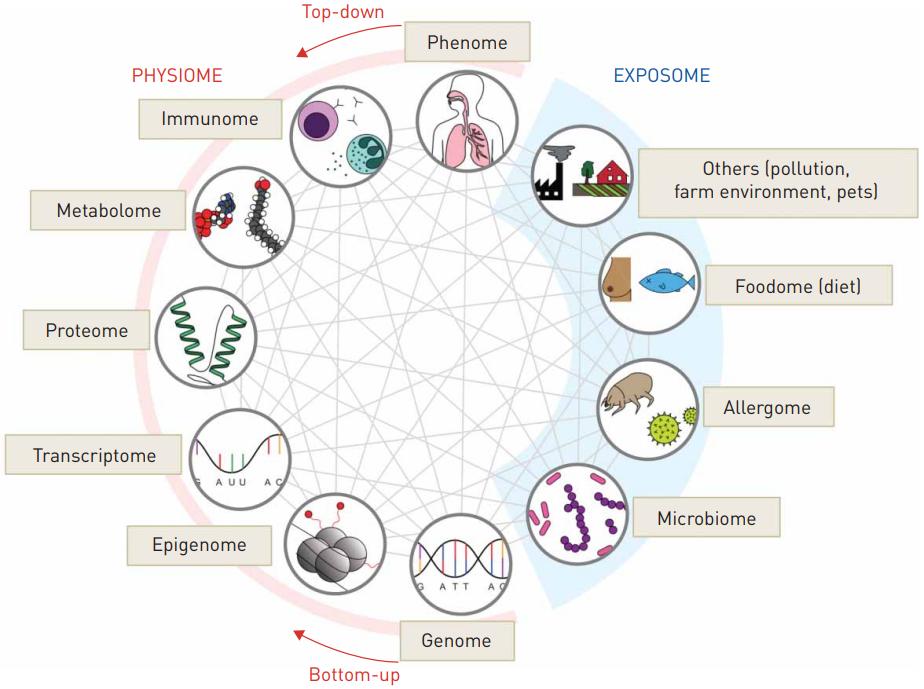 Fig.1 Omics approaches in allergy. (Tang, H. H., et al., 2020)
Fig.1 Omics approaches in allergy. (Tang, H. H., et al., 2020)
Genomics in Allergies
Immunomics has made a significant contribution to allergy research. Genomic approaches aim to identify genetic variants associated with specific phenotypes primarily through candidate gene studies and genome-wide association studies (GWAS).
A study based on GWAS has identified peanut allergy-specific loci in the human leukocyte antigen (HLA)-DR/-DQ region, which is part of the major histocompatibility complex loci involved in the expression of antigens to T cells. Functional annotation of allergy-related genes implies that these located motifs are involved in immune regulation and epidermal/epithelial barrier function, suggesting a role for related mechanisms in the etiology of FA.
Epigenomics in Allergies
Allergy is associated with genetics, environment, and genome-environment interactions, such as epigenetic effects. The epigenome lies at the intersection of environment, genotype, and cellular response.
A retrospective study has evaluated approximately 450,000 CpG in CD4+ T cells from 12 children diagnosed with IgE-mediated food allergy at 12 months of age. This study has identified 136 and 179 distinct methylation probes at birth and 12 months of age, respectively. Pathway analysis of these genes identified a number of mitogen-activated protein kinase signaling-associated molecules that may contribute to suboptimal early childhood T lymphocyte responses and the development of food allergy.
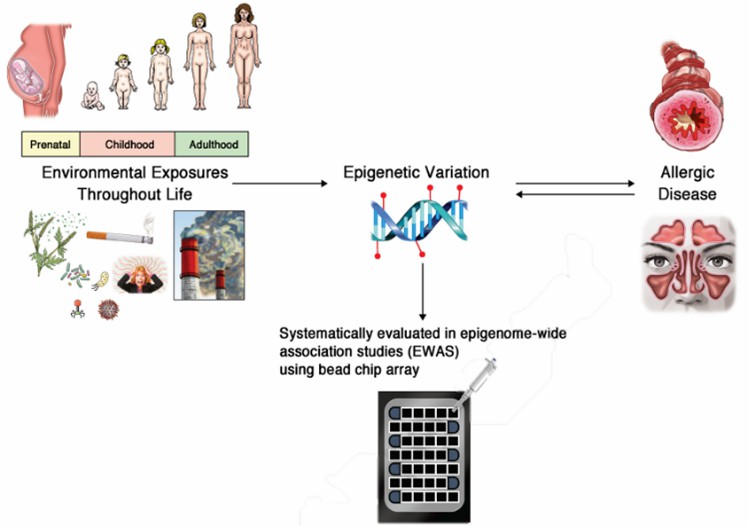 Fig.2 Interactions between environmental factors, epigenome, and allergic diseases. (Cardenas, A., et al., 2023)
Fig.2 Interactions between environmental factors, epigenome, and allergic diseases. (Cardenas, A., et al., 2023)
Transcriptomics in Allergies
Transcriptomic studies of allergies have facilitated the discovery of biomarkers of allergic reactions that can be used as diagnostic tools to monitor disease progression, select the most effective treatments, and help predict treatment outcomes.
For example, to characterize wheat allergens, screening the cDNA expression library of the wheat transcriptome with IgE antibodies from wheat allergic patients revealed that the high molecular weight glutenin subunit Bx7 allergen contains duplicated IgE epitopes. In addition, cellular-level transcriptomic studies are beginning to explore the general immune system and the involvement of TH1, TH2, and TH17 T cells in the pathogenesis of allergies.
Proteomics in Allergies
Proteomics is an important approach to the study of allergic reactions and is often used to identify novel biomarkers by studying human serum samples. The discovery of these novel markers can be used to uncover new pathophysiological mechanisms, differentiate between different allergy types, and turn into new therapeutic targets.
Recently, CyTOF has increasingly flourished in food allergies research as an advanced and in-depth proteomics technique. This technique facilitates high-dimensional quantitative analysis of cell populations and functional states at cellular-level resolution. The use of CyTOF to characterize immune cells and specific responses has been recently reported in several food allergy cohorts.
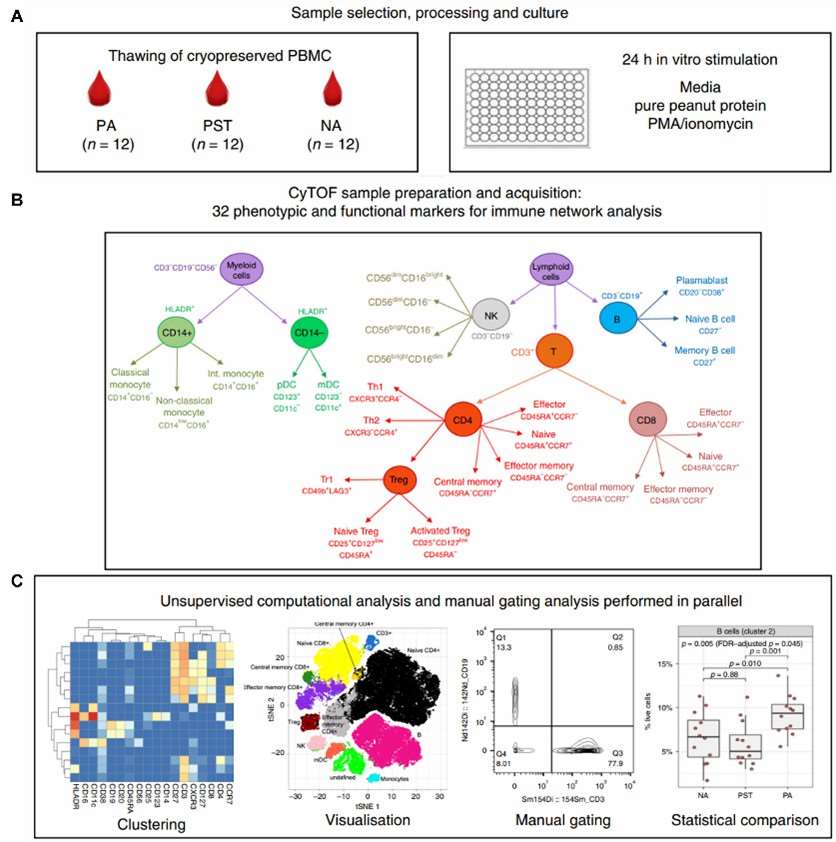 Fig.3 Experimental workflow for the mass spectrometric analysis of PBMCs. (Neeland, M. R., et al., 2020)
Fig.3 Experimental workflow for the mass spectrometric analysis of PBMCs. (Neeland, M. R., et al., 2020)
CD Genomics has a professional immunomics research team and equipment. We offer a wide range of immunomics-related services, including immunogenomics analysis, immune cell epigenomics analysis, immune cell transcriptomics analysis, immune cell proteomics analysis, and immunomics bioinformatics services to help clients understand the mechanisms of the immune system and discover novel drug and vaccines.
References
- Tang, H. H., Sly, P. D., Holt, P. G., Holt, K. E., & Inouye, M. (2020). Systems biology and big data in asthma and allergy: recent discoveries and emerging challenges. European Respiratory Journal, 55(1).
- Cardenas, A., Fadadu, R. P., & Koppelman, G. H. (2023). Epigenome-Wide Association Studies of Allergic Disease and the Environment. Journal of Allergy and Clinical Immunology.
- Neeland, M. R., Andorf, S., Manohar, M., Dunham, D., Lyu, S. C., Dang, T. D., ... & Nadeau, K. C. (2020). Mass cytometry reveals cellular fingerprint associated with IgE+ peanut tolerance and allergy in early life. Nature communications, 11(1), 1091.
